|
Protein Engineering on Leucyl-tRNA Synthetase (LeuRS)
Each of the twenty tRNA systhetases (RS) is
responsible for covalently linking a particular amino acid to its
cognate tRNA. Through a two-step
reaction, the enzyme forms an aminoacyl adenylate (AMP) and then
transfers the activated amino acid to the 3' end of the tRNA. The
'charged' tRNA
shuttles the amino acid to the directing protein systhesis.
Although the reaction is highly specific, some RS can activate and
aminoacylate
incorrect amino acids which structurally overlap with the cognate amino
acid. High fidelity of these RS is maintained via an extra domain of
about 200-300 amino acids containing a hydrolytic active site
that
proofreads and edits misactivated or mischarged aminoacids. Our goal is 'Protein Engineering' or
'Protein Design'
by controling of tRNA synthetase. By mutation of some proper
residue(s),
we expect to be able to control the amino-acid recognition ability(or
characteristics)
of a tRNA synthetase. In order to achieve this goal, we
chose
the leucyl tRNA synthetase system as a prove by the relationship with
cowork
group (Martinis
Group, University of Houston). Unfortunately, the 3D structure of E.
coli LeuRS is not known yet. Therefore we built the homology
modeling structure of E. coli LeuRS using the X-ray structure
of T. thermophilus LeuRS (K.W. Lee et al. Proteins 54(4)
693-704 (2004)). 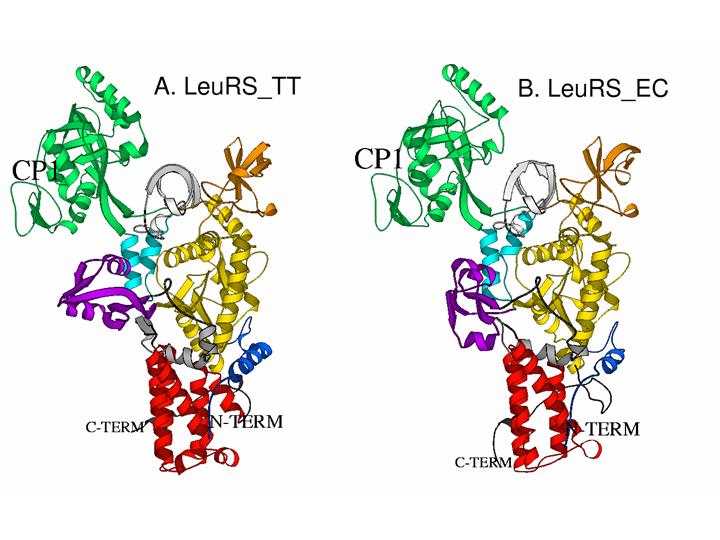
Fig.1. X-ray structures of T. Thermophilus LeuRS (A) and homology modeled structure of E. Coli LeuRS(B) From the refine
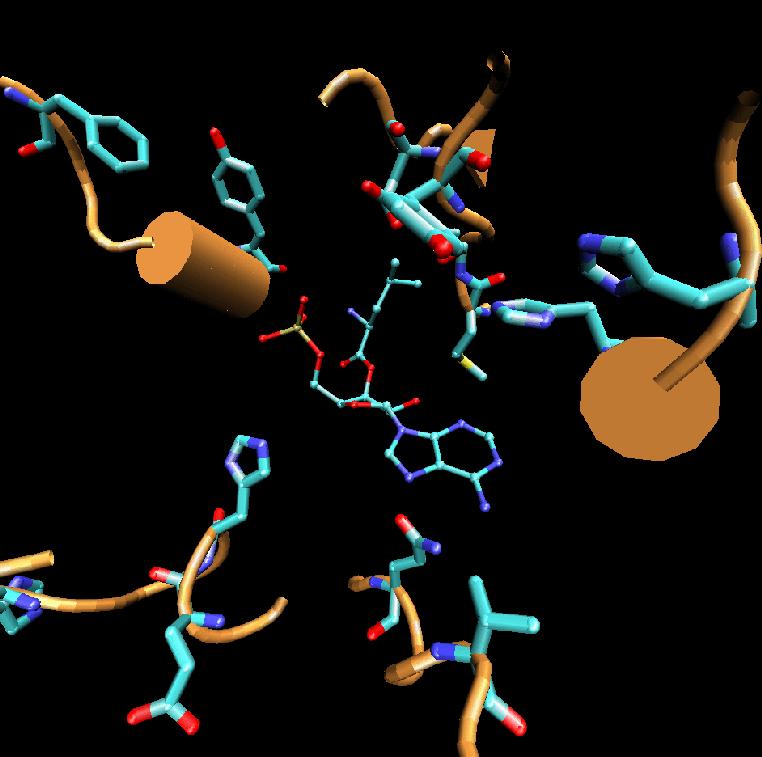
3D structure, the binding mode of the ligand leucine
and LeuRS was studied by the DOCKING simulation. Based on the
information from DOCKING result, the mutation residue will be decided
and then at the same time, many molecular dynamics (MD) simulations
were followed to investigate the interaction between the ligand and the
protein.
|