|
Membrane Porin Protein: OmpF (OmpK36, OmpC and PhoE)
Porins are integral outer-membrane proteins of Gram-negative bacteria that function as molecular sieves (with exclusion limits of around 600 Da) and are responsible for the uptake of small molecular nutrients and the removal of waste products. Since porin exists only in (gram negative) bacteria, it is the ideal target of antibacterial agents.
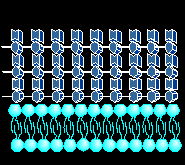
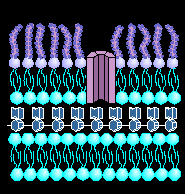
THE BACTERIAL CELL WALL STRUCTURE
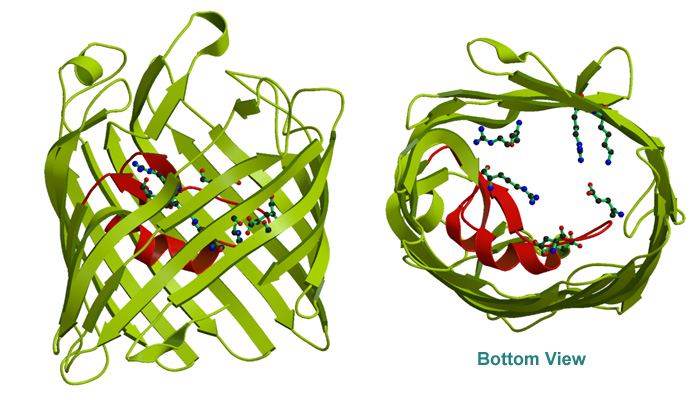
X-ray stucture have revealed a large
16-stranded antiparallel
barrel
structure enclosing the transmembrane pore. Three subunits form a
timmer. The 3-fold axis is approximately parallel to the barrel axis.
1.
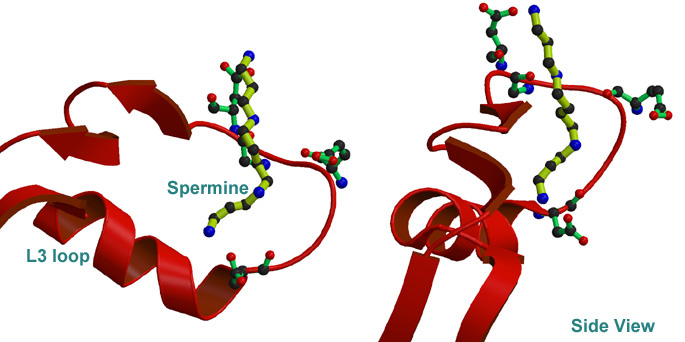
Electrostatics of porin pore region (i.e. L3 Loop).
2. Finding binding mode of polyamine to porin by Docking simulation 3. Dynamics of L3 loop of normal and ion(charge) induced status by MD simulation.
|
[More Porin Information from Paul's Homepage at Davidson College]